Pseudomonas aeruginosa
Pseudomonas aeruginosa is a is a highly virulent and multidrug resistant bacterium, primarily targeting individuals with reduced immunity. It was firstly described, isolated and recognised as a pathogen in the end of 19th century. P. aeruginosa causes between 10% and 20% of all infections.[1] It is on the ‘critical’ category list of the World Health Organisation (WHO) for research and developing the new drugs.[2]
| Pseudomonas aeruginosa | |
|---|---|
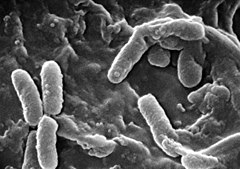
| |
| Scanning electron micrograph of Pseudomonas aeruginosa | |
| Scientific classification | |
| Kingdom: | |
| Phylum: | |
| Class: | |
| Order: | |
| Family: | |
| Genus: | |
| Species: | P. aeruginosa
|
| Binomial name | |
| Pseudomonas aeruginosa (Schröter 1872)
Migula 1900 | |

Biology
changeP. aeruginosa is a Gram-negative, rod-shaped bacterium, known by its environmental abundance. It can be found in water, soil, animal, human and plant environments, which can be explained by its versatile energy metabolism; it can survive without oxygen (anaerobic).[3] It has multiple hair-like attachments (pili) which are found on the surface of many bacteria and archaea to attach to other cells, and one flagellum, which is another but longer hair-like structure which provide motility.
Pathogenesis
changeP. aeruginosa primarily causes clinical infections in individuals with compromised immune systems. For example, in patients with cystic fibrosis. These people build up mucus layers, which create an environment for the bacteria that somewhat protects them from antibiotics and the host immune system. People with cancer, large wounds, and individuals on immunosuppressant drugs after transplant surgery are also considered to be susceptible.
P. aeruginosa may cause different infections, depending on the organ affected. Infections may include: pneumonia, diarrhoea, meningitis, urinary tract infections, bloodstream, ear and wound infections.[4][5] It is spread through improper hygiene, water contamination and medical equipment which is not properly sterilised. Along with high predisposition of patients with weak immunity, infections can be easily acquired in hospital, through, for example, medical devices, such as breathing machine and catheters.[4]
Antibiotic resistance and research
changeP. aeruginosa is a Gram-negative bacteria, containing two lipid bilayers (acting as barriers) with thin peptidoglycan layer (composed of proteins) in the middle. Because its peptidoglycan is covered by a lipid bilayer, some of the antibiotics, such as penicillin are less effective, as they target proteins within peptidoglycan.
P. aeruginosa also contains small circular genetic material (plasmids) which carry antibiotic resistance genes. These genes make proteins which will protect bacteria from antibiotics. Some of these proteins in P. aeruginosa are efflux pumps - transport proteins which pump the antibiotic outside of cells. Efflux pumps were firstly described in P. aeruginosa as well.
Resistant genes can be acquired through gene transfers. In vertical gene transfer, the new gene appears, for example, through spontaneous mutation. In horizontal gene transfer, the gene is transferred from one bacterium to another. This type of gene transfer makes most of the evolution and determines the bacterium virulence and antibiotic resistance.[6][7]
Recent research shows that phage therapy (involving bacteriophages – bacterial viruses) might be considered as a possible treatment. Bacteriophages attack and destroy extracellular matrix (cell skeleton), increasing cell permeability to antibiotics.[8]
Diagnosis and treatment
changeP. aeruginosa diagnosis includes physical examination, along with fluid samples. Mild infections can be treated with certain antibiotics. It becomes difficult for the severe cases due to antibiotic resistance. Often the samples are taken into laboratory to see which antibiotics are effective for the strain.
References
change- ↑ Anderson, T. R.; Slotkin, T. A. (1975-08-15). "Maturation of the adrenal medulla--IV. Effects of morphine". Biochemical Pharmacology. 24 (16): 1469–1474. doi:10.1016/0006-2952(75)90020-9. ISSN 1873-2968. PMID 7.
- ↑ Makar, A. B.; McMartin, K. E.; Palese, M.; Tephly, T. R. (June 1975). "Formate assay in body fluids: application in methanol poisoning". Biochemical Medicine. 13 (2): 117–126. doi:10.1016/0006-2944(75)90147-7. ISSN 0006-2944. PMID 1.
- ↑ Encyclopedia of food microbiology. Carl A. Batt, Mary Lou Tortorello (2nd ed.). Amsterdam. 2014. ISBN 978-0-12-384733-1. OCLC 881482027.
{{cite book}}: CS1 maint: location missing publisher (link) CS1 maint: others (link) - ↑ 4.0 4.1 Wiesmann, U. N.; DiDonato, S.; Herschkowitz, N. N. (1975-10-27). "Effect of chloroquine on cultured fibroblasts: release of lysosomal hydrolases and inhibition of their uptake". Biochemical and Biophysical Research Communications. 66 (4): 1338–1343. doi:10.1016/0006-291x(75)90506-9. ISSN 1090-2104. PMID 4.
- ↑ Bodey, G. P.; Bolivar, R.; Fainstein, V.; Jadeja, L. (March 1983). "Infections caused by Pseudomonas aeruginosa". Reviews of Infectious Diseases. 5 (2): 279–313. doi:10.1093/clinids/5.2.279. ISSN 0162-0886. PMID 6405475.
- ↑ Botelho, João; Grosso, Filipa; Peixe, Luísa (May 2019). "Antibiotic resistance in Pseudomonas aeruginosa - Mechanisms, epidemiology and evolution". Drug Resistance Updates: Reviews and Commentaries in Antimicrobial and Anticancer Chemotherapy. 44: 100640. doi:10.1016/j.drup.2019.07.002. ISSN 1532-2084. PMID 31492517. S2CID 199640645.
- ↑ Bose, K. S.; Sarma, R. H. (1975-10-27). "Delineation of the intimate details of the backbone conformation of pyridine nucleotide coenzymes in aqueous solution". Biochemical and Biophysical Research Communications. 66 (4): 1173–1179. doi:10.1016/0006-291x(75)90482-9. ISSN 1090-2104. PMID 2.
- ↑ Chegini, Zahra; Khoshbayan, Amin; Taati Moghadam, Majid; Farahani, Iman; Jazireian, Parham; Shariati, Aref (2020-09-30). "Bacteriophage therapy against Pseudomonas aeruginosa biofilms: a review". Annals of Clinical Microbiology and Antimicrobials. 19 (1): 45. doi:10.1186/s12941-020-00389-5. ISSN 1476-0711. PMC 7528332. PMID 32998720.