Simplified molecular input line entry specification
This article uses too much jargon, which needs explaining or simplifying. (January 2024) |
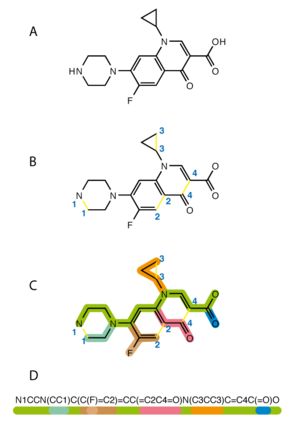
The simplified molecular input line entry specification or SMILES is a line notation for the chemical structure of molecules. It uses short series ASCII characters to represent structures. Most molecule editor computer programs can draw a two-dimensional diagram or a three-dimensional model of a molecule based on its SMILES code.
Arthur Weininger and David Weininger wrote the original SMILES specification in the late 1980s. It has since been modified and extended by others, including by Daylight Chemical Information Systems Inc. In 2007, the Blue Obelisk open-source chemistry community developed an open standard called "OpenSMILES".
In July 2006, the International Union of Pure and Applied Chemistry introduced the InChI as a standard line notation for representing chemical structures. People think that it is easier for humans to understand SMILES than InChI. Some chemists argue that SMILES is better than InChI because many different software programs support it. SMILES also has theoretical (for example, graph theory) backing.
Conversion
changeSMILES can be converted back to 2-dimensional representations using Structure Diagram Generation algorithms.[1] This conversion can sometimes be ambiguous. Because SMILES just says which atoms are connected without any information about the angles between chemical bonds, computer programs use energy minimization to convert SMILES to 3-dimensional representations. There are many downloadable and web-based conversion utilities.
Examples
changeHere are some examples of the SMILES for molecules:
| Molecule | Structure | SMILES Formula |
|---|---|---|
| Dinitrogen | N≡N | N#N |
| Methyl isocyanate (MIC) | CH3–N=C=O | CN=C=O |
| Copper(II) sulfate | Cu2+ SO42- | [Cu+2].[O-]S(=O)(=O)[O-] |
| Œnanthotoxin (C17H22O2) | CCC[C@@H](O)CC\C=C\C=C\C#CC#C\C=C\CO | |
| Pyrethrin II (C22H28O5) | COC(=O)C(\C)=C\C1C(C)(C)[C@H]1C(=O)O[C@@H]2C(C)=C(C(=O)C2)CC=CC=C | |
| Aflatoxin B1 (C17H12O6) | O1C=C[C@H]([C@H]1O2)c3c2cc(OC)c4c3OC(=O)C5=C4CCC(=O)5 | |
| Glucose (glucopyranose) (C6H12O6) | OC[C@@H](O1)[C@@H](O)[C@H](O)[C@@H](O)[C@@H](O)1 | |
| Bergenin (cuscutin) (a resin) (C14H16O9) | OC[C@@H](O1)[C@@H](O)[C@H](O)[C@@H]2[C@@H]1c3c(O)c(OC)c(O)cc3C(=O)O2 | |
| A pheromone of the Californian scale insect | CC(=O)OCCC(/C)=C\C[C@H](C(C)=C)CCC=C | |
| 2S,5R-Chalcogran: a pheromone of the bark beetle Pityogenes chalcographus[2] | CC[C@H](O1)CC[C@@]12CCCO2 | |
| Vanillin | O=Cc1ccc(O)c(OC)c1 | |
| Melatonin (C13H16N2O2) | CC(=O)NCCC1=CNc2c1cc(OC)cc2 | |
| Flavopereirin (C17H15N2) | CCc(c1)ccc2[n+]1ccc3c2Nc4c3cccc4 | |
| Nicotine (C10H14N2) | CN1CCC[C@H]1c2cccnc2 | |
| Alpha-thujone (C10H16O) | CC(C)[C@@]12C[C@@H]1[C@@H](C)C(=O)C2 | |
| Thiamin (C12H17N4OS+) (vitamine B1) |
OCCc1c(C)[n+](=cs1)Cc2cnc(C)nc(N)2 |
References
change- ↑ Helson, H.E. (1999). "Structure Diagram Generation". In Lipkowitz, K. B. and Boyd, D. B. (ed.). Rev. Comput. Chem. Reviews in Computational Chemistry. Vol. 13. New York: Wiley-VCH. pp. 313–398. doi:10.1002/9780470125908.ch6. ISBN 9780470125908.
{{cite book}}: CS1 maint: multiple names: editors list (link) - ↑ Isolation of Pheromone Synergists of Bark Bettle, Pityogenes chalcographus, From Complex Insect-Plant Odors by Fractionation and Subtractive-Combination Bioassay
- Anderson, E.; Veith, G.D; Weininger, D. (1987). SMILES: A line notation and computerized interpreter for chemical structures. Report No. EPA/600/M-87/021. Duluth, MN 55804: U.S. EPA, Environmental Research Laboratory-Duluth.
{{cite book}}: CS1 maint: location (link) CS1 maint: multiple names: authors list (link) - Weininger, David (1988). "SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules". Journal of Chemical Information and Modeling. 28: 31–36. doi:10.1021/ci00057a005. S2CID 5445756.
- Weininger, David; Weininger, Arthur; Weininger, Joseph L. (1989). "SMILES. 2. Algorithm for generation of unique SMILES notation". Journal of Chemical Information and Modeling. 29 (2): 97. doi:10.1021/ci00062a008. S2CID 6621315.
- Helson, H.E. (1999). "Structure Diagram Generation". In Lipkowitz, K. B. and Boyd, D. B. (ed.). Rev. Comput. Chem. Reviews in Computational Chemistry. Vol. 13. New York: Wiley-VCH. pp. 313–398. doi:10.1002/9780470125908.ch6. ISBN 9780470125908.
{{cite book}}: CS1 maint: multiple names: editors list (link)
Other websites
changeSpecifications
change- "SMILES - A Simplified Chemical Language"
- The OpenSMILES home page
- "SMARTS - SMILES Extension"
- Daylight SMILES tutorial
- Parsing SMILES
SMILES related software utilities
change- NCI/CADD Chemical Identifier Resolver – resolves or generates SMILES from chemical names, CAS Registry Numbers, InChI/InChIKey and many other chemical structure file formats
- NCI/CADD Online SMILES Translator and Structure File Generator Archived 2001-05-01 at the Wayback Machine – Java online molecule editor
- PubChem server side structure editor – online molecule editor
- smi23d Archived 2007-09-14 at the Wayback Machine – 3D Coordinate Generation
- Daylight Depict Archived 2001-12-02 at the Wayback Machine – Translate a SMILES formula into graphics
- GIF/PNG-Creator for 2D Plots of Chemical Structures Archived 2004-10-15 at the Wayback Machine
- JME molecule editor Archived 2001-04-28 at the Wayback Machine
- ACD/ChemSketch Archived 2006-10-18 at the Wayback Machine
- Marvin Archived 2007-11-07 at the Wayback Machine by ChemAxon – online chemical editor/viewer and SMILES generator/converter
- Instant JChem Archived 2007-11-12 at the Wayback Machine by ChemAxon – desktop application for storing/generating/converting/visualizing/searching SMILES structures, particularly batch processing; personal edition free
- JChem for Excel Archived 2010-02-03 at the Wayback Machine by ChemAxon – MS Excel add-in for storing/generating/converting/visualizing/searching SMILES structures
- Smormo-Ed – a molecule editor for Linux which can read and write SMILES
- InChI.info – an unofficial InChI website featuring on-line converter from InChI and SMILES to molecular drawings
- Balloon – A free program for 3D coordinate generation and conformational analysis.
- Indigo Archived 2015-02-11 at the Wayback Machine – an open-source cross-platform cheminformatics library with a plugin for IUPAC-compliant molecule and reaction 2D structural formula rendering.
- Open Babel – an open-source chemical toolbox allowing anyone to search, convert, analyze, or store biochemical data.
- Bioclipse Archived 2010-01-11 at the Wayback Machine – a free and open source workbench for the life sciences
- MolEngine Archived 2019-09-15 at the Wayback Machine – A .NET cheminformatics toolkit to read/write SMILES, generate 2D coordinate from SMILES, and convert SMILES from/into other Chemical file formats.
- JSDraw Archived 2011-07-17 at the Wayback Machine – A cross-platform javascript chemical structure editor to generate SMILES and SMARTS.